Climate change is exacerbating water scarcity and negatively impacting water quality through heavy rain events and droughts in many regions of the world. A better understanding of the spread of microbiological contaminants in groundwater is important to counteract this.
The DFG funded project PrePat ("Development and application of non-pathogens and extracellular DNA for predicting transport and attenuation of pathogens and antibiotic resistance genes in groundwater") aims to improve the understanding of the transport of bacteria, viruses and antibiotic resistance genes in groundwater.
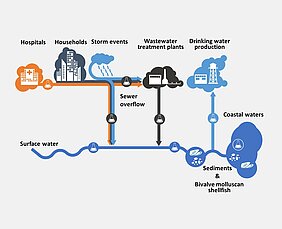
Karst aquifers are susceptible to contamination because of their special properties, which allow rapid groundwater recharge and high groundwater velocities in the saturated zone. Microbiological contaminants such as bacteria and viruses can enter the groundwater and reach the springs, for example through the fertilisation of agricultural land.
In the well-studied catchment area of the Gallus spring (Swabian Alb, south-west Germany), rainfall events occasionally lead to overflows from a rainwater retention basin, causing wastewater to infiltrate into the groundwater. After these overflows, bacteria and organic trace substances can be detected in the Gallus spring, which is used for drinking water production and is about 9 km away. When analysing these overflows, there is a lack of information on the time of entry and the concentration of the pollutants, which limits the determination of their transport and retention parameters.
As part of PrePaT, tracer tests were carried out with various non-pathogenic bacteria, bacteriophages and extracellular DNA (eDNA) together with uranine in the catchment. The average flow velocities in the groundwater system during the tracer tests were 30 and 87 m/s. The tracers were injected as Dirac injections within 10 minutes and regular sampling for the biological tracers started with the breakthrough of uranine approximately 80 to 90 hours after injection. Bacterial and eDNA tracers were analysed using qPCR methods, while bacteriophages were additionally analysed using a culture-based method (plaque assay) to quantify active phages.
All tracers were successfully injected into the groundwater. The results of the first tracer test showed that the tracers used were transported at least 3 km within the system. In addition, the active bacteriophages from the second tracer test were recovered within 90 hours after being transported 9 km from the stormwater retention basin to the source. Results for microbiological parameters analysed using PCR-based methods were mostly below the limit of quantification at the source. Effects such as increased retention of phages, bacteria and eDNA, their degradation or loss of sample material due to the need for sample enrichment and nucleic acid extraction prior to PCR analysis can further reduce the theoretical maximum concentration and lead to concentrations close to or below the limit of quantification of the PCR method used. This is where culture methods for bacteriophage detection, with their higher sensitivity, offer an advantage. Aquabacterium citratiphilum B4 was used, a bacterium for which no specific culture method is available.
The modelling showed significantly lower recovery rates for the active phages than for the soluble tracer uranine (maximum ~1% compared to ~31% for uranine). The breakthrough of infectious bacteriophages at the source within a few days shows the high contamination potential of karst springs even with relatively low input masses and long transport distances. Both bacteriophages used showed comparable breakthrough curves with the conventional soluble tracer, indicating similar transport processes.
Publication
Stange, C.; Tiehm, A.: Occurrence of antibiotic resistance genes and microbial source tracking markers in the water of a karst spring in Germany. The Science of the total environment 742: 140529 (2020) DOI:/10.1016/j.scitotenv.2020.140529
Schiperski, F., Zirlewagen, J., Stange, C., Tiehm, A., Licha, T., Scheytt, T.: Transport-based source tracking of contaminants in a karst aquifer: Model implementation, proof of concept, and application to event-based field data. Water research 213, 118145 (2022). DOI:/10.1016/j.watres.2022.118145
Serbe, R.; Schiperski, F.; Stelmaszyk, L.; Stange, C.; Tiehm, A.; Scheytt, T.: Long-distance transport of bacteriophages MS2 and phiX174 in karst aquifers. Science of the Total Environment 973, 179111 (2025) DOI:/10.1016/j.scitotenv.2025.179111
Serbe, R., Schiperski, F., Stelmaszyk, L., Stange, C., Scheytt, T.: Transport of bacteria, bacteriophages, and extracellular DNA as surrogates for pathogens and antibiotic resistance genes in a karst aquifer (Gallusquelle, South-West Germany), EGU23, the 25th EGU General Assembly, held 23-28 April, 2023 in Vienna, Austria and Online. Online at egu23.eu, id. EGU-11903. ui.adsabs.harvard.edu/link_gateway/2023EGUGA..2511903S/doi:10.5194/egusphere-egu23-11903

![[Translate to English:] Prüfstelle-Produktprüfung_Teststand Test centre and product testing](/fileadmin/_processed_/0/9/csm_TZW-Karlsruhe_Pruefung_Geraete-Teststand_377188946c.jpg)