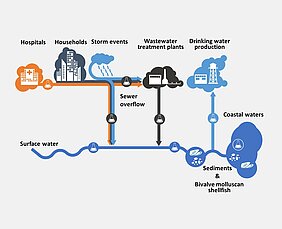
A better understanding of the fate of pathogenic viruses and antibiotic-resistant bacteria from the sources to river basins and estuaries are urgently required. The international SARA research project, coordinated by TZW, was able to close a number of knowledge gaps.
Our project will determined the prevalence of pathogenic viruses (including SARS-CoV-2), microbial indicators, antibiotic resistance, and microbial source tracking (MST) markers in wastewater, surface water, coastal sea waters, sediment and bivalve molluscan shellfish (BMS) in catchments located in different climatic zones (Sweden, Germany, France, Spain, Portugal, Israel, Mozambique, and Uganda). The project aims are: (i) method harmonization and training of European and African partners, (ii) detection of SARS-CoV-2 detection in raw wastewater as a biomarker for COVID-19 cases, (iii) enteric viruses, antibiotic resistances and MST markers monitoring in aquatic environments, (iv) evaluation of sediments and BMS as integral reservoirs, (v) determination of the impact of climate and extreme weather events, and (vi) microbial risk assessment for water resources.
The project was accompanied by a Stakeholder Forum consisting of representatives of international health and environmental organizations as well as authorities and water utilities.
Culture, molecular biological and metagenomic methods were used for testing. Culture-based analyses were performed in all partner laboratories within 24 or 48 hours. For the molecular analyses, samples were immediately enriched, extracted and the resulting nucleic acid extracts stored at -80°C until sent to the specialised partner laboratory. To ensure comparability of results, these methods were harmonised in the first phase of the project and summarised in a brochure (available on the SARA project website).
Particular attention was paid to capacity building of the African partners. Following the successful harmonisation of methods, the European partners helped to install the laboratory equipment in the African partner laboratories and provided extensive training to the African laboratory staff. The training included the harmonised methods for culture detection of indicator microorganisms and antibiotic resistant bacteria, as well as the initial concentration and extraction steps of samples for molecular biological analysis.
Following the successful harmonisation of methods and selection of key antibiotic resistance genes, sampling campaigns were carried out at the different model sites. Overall, the SARA project provided important insights into the prevalence of pathogenic viruses, microbial indicators, antibiotic resistance and MST markers in wastewater, surface water, coastal seawater, sediment and BMS in catchments in different climate zones.
The project results show:
- the advantages of wastewater-based surveillance for SARS-CoV-2
- differences in the presence of the investigated human pathogenic viruses between the different catchments and wastewater treatment plants
- differences in the prevalence and relative abundance of Extended spectrum beta-lactamase (ESBL)-producing E. coli and antibiotic resistance genes between the model sites, with the highest abundance in Uganda and the lowest abundance in Sweden
- higher levels of somatic coliphages as viral indicators compared to F-specific bacteriophages
- limited reduction for all microbiological parameters studied in the Ugandan stabilisation ponds in their current mode of operation
- an increase in microbial contamination following extreme events (heavy rainfall)
- high persistence of antibiotic resistant environmental oligotrophic bacteria as an important reservoir of antimicrobial resistance in the environment
- presence of all three human MST markers detected, indicating high levels of human faecal contamination at all model sites.
- increasing evidence that bacteriophages play an important role in the spread of antibiotic resistance genes in aquatic ecosystems.
The most important findings from the project were summarised in a document that can be viewed on the SARA project website.
Publications
Abaasa C. N., Stange C., Ayesiga S., Mulogo E. M., Rogers K., Lejju J. B., Andama M., Tamwesigire I. K., Bazira J., Byarugaba F., Tiehm A. Antibiotic resistance of E. coli isolates form different water sources in Mbarara, Uganda. J. Water Health (2024) DOI: 10.2166/wh.2024.319
Holzer, C.; Ho, J.; Tiehm, A.; Stange, C.: Wastewater monitoring - passive sampling for the detection of SARS-CoV-2 and antibiotic resistance genes in wastewater. The Science of the Total Environment 959, 178244 (2025) DOI: 10.1016/j.scitotenv.2024.178244
Stange C.,Sidhu J.P.S.,Tiehm A.,Toze S.: Antibiotic resistance and virulence genes in coliform water isolates.
International Journal of Hygiene and Environmental Health 219: 823-831 (2016) DOI.org/10.1016/j.ijheh.2016.07.015
Stange, C.; Yin, D.; Xu, T.; Guo, X.; Schäfer, C.; Tiehm, A.: Distribution of clinically relevant antibiotic resistance genes in Lake Tai, China. Science of the total environment 655: 337–346 (2019) DOI: 10.1016/j.scitotenv.2018.11.211
Ho J., Seidel M., Niessner R., Eggert J., Tiehm A.: Long amplicon (LA)-qPCR for the discrimination of infectious and noninfectious phiX174 bacteriophages after UV inactivation. Water Research 103: 141-148 (2016) DOI 10.1016/j.watres.2016.07.032
Poster
Stange C., Ho J., Sanchez-Cid C., Mulogo E., Nasser A., Nhantumbo C., Monteiro S., Simonsson M., Blanch A.R., Vogel T.M, Tiehm A.: Wastewater-based epidemiology for monitoring COVID-19 and antimicrobial resistance: Insights from comprehensive studies. International Conference Towards a Global Wastewater Surveillance System for Public Health, 15-17 Nov 2023, Frankfurt am Main, Germany (pdf-Datei)

![[Translate to English:] Prüfstelle-Produktprüfung_Teststand Test centre and product testing](/fileadmin/_processed_/0/9/csm_TZW-Karlsruhe_Pruefung_Geraete-Teststand_377188946c.jpg)
























![[Translate to English:]](/fileadmin/_processed_/8/0/csm_Bild_SARA_DE_0c92e7a2be.jpg)