The increase in antibiotic resistance has been termed a "silent pandemic" by the World Health Organization (WHO). The long-standing and extensive use of antibiotics has favoured the formation and spread of antibiotic-resistant bacteria and antibiotic resistance genes not only in the clinical setting but also in the aquatic environment. For this reason, antibiotic-resistant bacteria and antibiotic resistance genes are increasingly discussed as new parameters for assessing hygienic water quality. TZW has many years of experience in the detection of antibiotic resistance in the environment through a number of completed and ongoing research projects. In this article we present a recently completed project and its results.
Within the framework of a project funded by DVGW over a period of three and a half years, adapted methods for the detection of antibiotic-resistant bacteria in the aquatic environment and of resistance genes after drinking water treatment and disinfection were developed and applied.
Detection of resistant environmental bacteria
Antibiotic resistance and traces of antibiotics enter the aquatic environment via wastewater or agriculture, e.g. through fertilization with manure from animal farming. Long-term inputs have promoted the spread of resistance among environmental bacteria adapted to the nutrient-poor conditions of surface waters. Of particular relevance are antibiotic resistances to clinical reserve antibiotics, which are classified as critical by the WHO. Reserve antibiotics are special antibiotics that are only used for infections with pathogens that are already resistant to common antibiotics. New culture methods have been developed that allow specific monitoring of resistant environmental bacteria. It has been shown that resistance not only to commonly applied antibiotics but also to reserve antibiotics is already widespread among environmental bacteria. A reduction of inputs at the sources is urgently needed.
Detection of resistance genes after treatment and disinfection
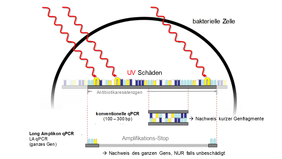
Reactive processes such as UV irradiation, chlorination and ozonation are common techniques in drinking water treatment. They lead to DNA damage and can therefore inactivate resistant bacteria and resistance genes. Antibiotic resistance genes have a size of up to 2000 base pairs and must be present as an intact gene to confer resistance. For the detection of resistance genes in standard molecular biology methods (qPCR), only small sections of these genes (100 to 300 base pairs) are examined. If exactly these subsections are not damaged after reactive treatments, false-positive results may occur. In order to realistically detect elimination efficiencies, so-called long amplicon methods have been successfully developed to detect the complete resistance genes. It could be shown in lab experiments and samples from drinking water treatment that the elimination performance of the three reactive methods is underestimated by up to a factor of 90 with the standard method compared to the long amplicon method.
Conclusion and outlook
The results of the project confirm the relevance of environmental bacteria as a reservoir for antibiotic resistance and offer the possibility of optimized detection after reactive water treatment. The detection and elimination of antibiotic resistances are currently being further developed and comparatively investigated in projects for the advanced treatment and reuse of wastewater ("reclaimed water", German title “Nutzwasser”) and in an international cooperation with eight partners in Europe and Africa ("SARA - Surveillance of Emerging Pathogens and Antibiotic Resistances in Aquatic Ecosystems").
Links
Publications
Stange C., Tiehm A.: Verhalten von Antibiotika-Resistenzgenen bei der Trinkwasseraufbereitung. Veröffentlichungen aus dem Technologiezentrum Wasser, ISSN 1434-5765, TZW-Band 76 (2017)
Stange C.,Sidhu J.P.S.,Tiehm A.,Toze S.: Antibiotic resistance and virulence genes in coliform water isolates. International Journal of Hygiene and Environmental Health 219: 823-831 (2016) DOI 10.1016/j.ijheh.2016.07.015
Stoll C., Sidhu J.P.S., Tiehm A., Toze S.: Prevalence of clinically relevant antibiotic resistance genes in surface water samples collected from Germany and Australia. Environmental Science & Technology 46: 9716-9726 (2012) DOI.org/10.1021/es302020s
Stange C., Sidhu J.P.S., Toze S., Tiehm A.: Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 222: 541-548 (2019) DOI 10.1016/j.ijhe.2019.02.002
Stange C., Yin D., Xu T., Guo X., Schäfer C., Tiehm A.: Distribution of clinically relevant antibiotic resistance genes in Lake Tai, China. Science of the Total Environment 655: 337-346 (2019) DOI 10.1016/j.scitotenv.2018.11.211
Stange, C.; Tiehm, A. (2020): Occurrence of antibiotic resistance genes and microbial source tracking markers in the water of a karst spring in Germany. The Science of the total environment 742, S. 140529. DOI: 10.1016/j.scitotenv.2020.140529.

![[Translate to English:] Prüfstelle-Produktprüfung_Teststand Test centre and product testing](/fileadmin/_processed_/0/9/csm_TZW-Karlsruhe_Pruefung_Geraete-Teststand_377188946c.jpg)